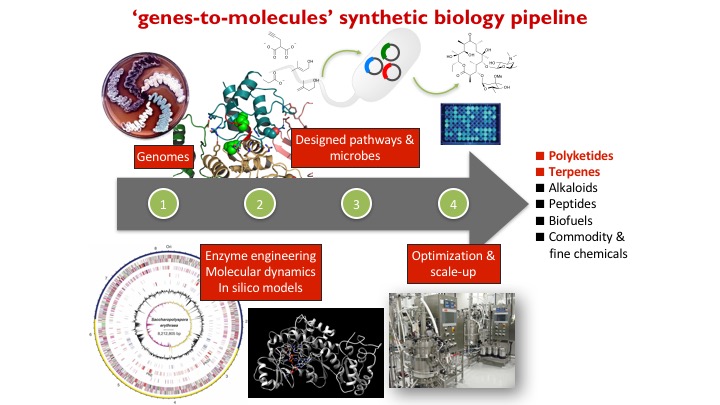
Overview
We are developing a genes-to-molecules synthetic biology pipeline for the synthesis, diversification, and discovery of natural products for drug discovery. We are interested in (1) how natural products are biosynthesized in Nature, (2) how to reprogram natural product biosynthesis to make new analogues, and (3) structure-function relationship studies of biologically active natural products.
Protein engineering and synthetic biology for polyketide diversification
Polyketides are a large group of plant and microbial secondary metabolites (natural products) from which many blockbuster drugs and agricultural/animal health products are derived, including antibiotics (erythromycin), anticancer drugs (doxorubicin), insecticides (spinosyn), anti-parasitics (avermectin), anti-fungals (amphotericin), anti-cholesterol (lovastatin), and immunosuppresants (rapamycin). Polyketides are biosynthesized by huge enzymatic assemblies whereby enzyme activities are organized into modules. Each module is responsible for the selection and condensation of small molecule building blocks into the growing product chain. Thus, the structure of the natural product is determined by the number, specificity, and order of modules within the assembly line, raising the exciting prospect of designing new hybrid biosynthetic systems for the synthesis of designer molecules.
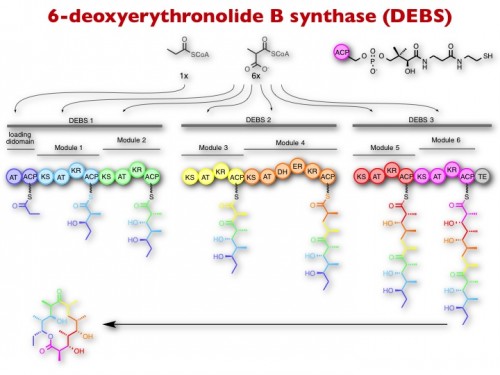 We are learning how to make such hybrid assembly lines and exploring strategies to install non-natural building blocks into polyketides by engineering the substrate specificity of individual modules and domains. To facilitate this process, we are also interested in developing and leveraging genome engineering methods based on CRISPR-Cas.
We are learning how to make such hybrid assembly lines and exploring strategies to install non-natural building blocks into polyketides by engineering the substrate specificity of individual modules and domains. To facilitate this process, we are also interested in developing and leveraging genome engineering methods based on CRISPR-Cas.
Biosensor-guided engineering of natural product biosynthesis
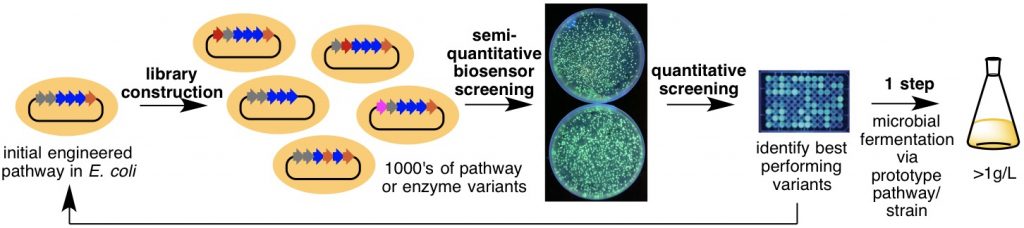 Often, the ability to access analogues of natural products for drug discovery and medicinal chemistry are restricted by our limited ability to manipulate natural product biosynthesis in microbes. For example, the large size and staggering complexity of polyketide biosynthesis renders the rational redesign and optimization of such pathways for improved yield and generation of analogues difficult, if not impossible in some cases. To address this major limitation, we have created genetically encoded biosensors that produce a fluorescent signal in the presence of the target natural product. These designer biosensors enable ultra-high throughput approaches to engineering natural product biosynthesis, and are being used to screen the productivity of thousands of pathway variants, thus bypassing our limited ability to rationally reprogram complex biosynthetic pathways.
Often, the ability to access analogues of natural products for drug discovery and medicinal chemistry are restricted by our limited ability to manipulate natural product biosynthesis in microbes. For example, the large size and staggering complexity of polyketide biosynthesis renders the rational redesign and optimization of such pathways for improved yield and generation of analogues difficult, if not impossible in some cases. To address this major limitation, we have created genetically encoded biosensors that produce a fluorescent signal in the presence of the target natural product. These designer biosensors enable ultra-high throughput approaches to engineering natural product biosynthesis, and are being used to screen the productivity of thousands of pathway variants, thus bypassing our limited ability to rationally reprogram complex biosynthetic pathways.
Synthetic biology of isoprenoid biosynthesis
We have developed an artificial two-enzyme pathway to provide access to natural and new-to-nature isoprenoids. The pathway is being coupled to various downstream biosynthetic steps to afford isoprenoids relevant to medicinal chemistry and the biotechnology industry. The simplicity and modularity of this system are being leveraged by directed evolution to expand its synthetic capabilities.
Probing the promiscuity and specificity of biosynthetic machinery
Biosynthetic enzymes usually display very narrow substrate specificities and are not useful for diversification of natural products. We have identified several enzymes from the biosynthesis of antibiotics that are promiscuous towards dozens of non-natural building blocks. The molecular basis for broad substrate specificity is being elucidated. In addition, we are investigating the molecular basis for regioselective installation of building blocks into natural products by probing and manipulating protein:protein interactions between various biosynthetic components.
Drug discovery and chemical biology
We are applying our gene-to-molecules synthetic biology pipeline to the diversification of antibiotics and anti-cancer scaffolds. Libraries of diversified natural products will be screened for new and useful biological activities or pharmacological properties.