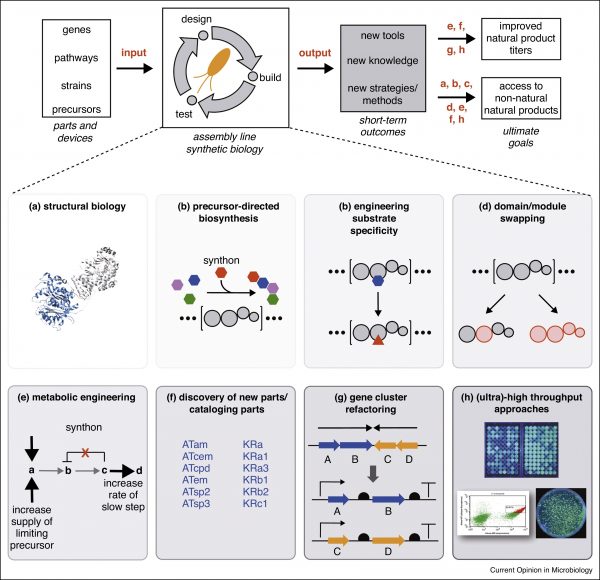
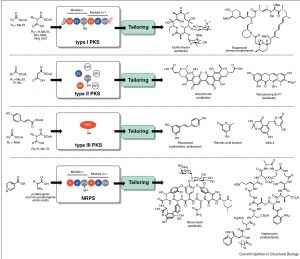
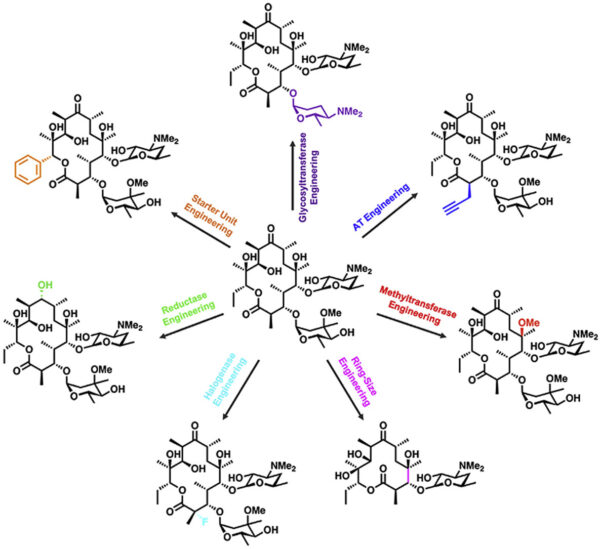
[54] Paulsel T, Williams GJ*. Current State-of-the-Art Toward Chemoenzymatic Synthesis of Polyketide Natural Products, 2023, ChemBioChem, OAP.
Very Important Paper, ChemBioChem
Editors Choice Spotlight at ChemEurope
[53] Welch S, Cossin J, Paulsel T, Cropp A, Williams GJ*. “Targeted enzyme modifications enable regioselective biosynthesis of fluorinated polyketides”, 2022, Chem Catalysis, 2, 10, 2440.
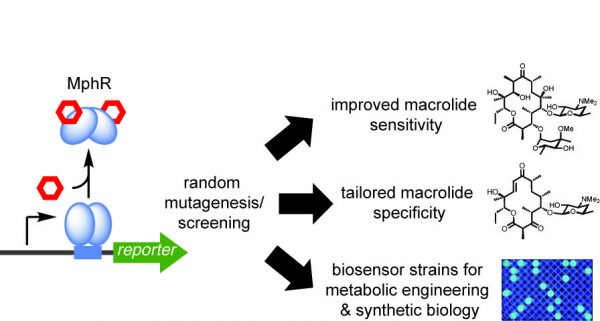
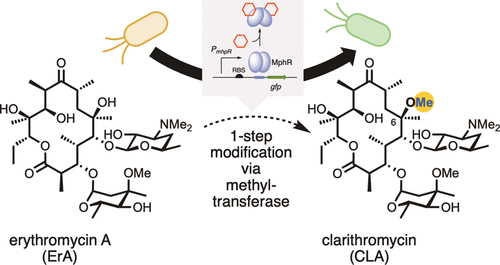
[52] Li Y, Reed M, Wright T, Cropp A, Williams GJ*. “Development of a genetically-encoded biosensor for reporting the methyltransferase-dependent biosynthesis of semi-synthetic macrolide antibiotics”, 2021, ACS Synth Biol, 10, 10, 2520.
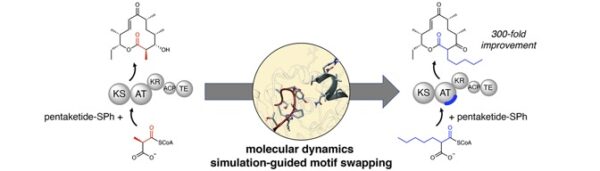
[51] Kalkreuter E, Bingham K, Keeler A, Lowell A, Schmidt J, Sherman DH, Williams GJ*. “Computationally-guided exchange of substrate selectivity motifs in a modular polyketide synthase acyltransferase”, 2021, Nat Commun, 12, 2193.
[50] Mitchler MM, Garcia JM, Montero NE, Williams GJ*. “Transcription factor-based biosensors: a molecular-guided approach for natural product engineering“. 2021, Curr Opin Biotechnol, 69, 172.
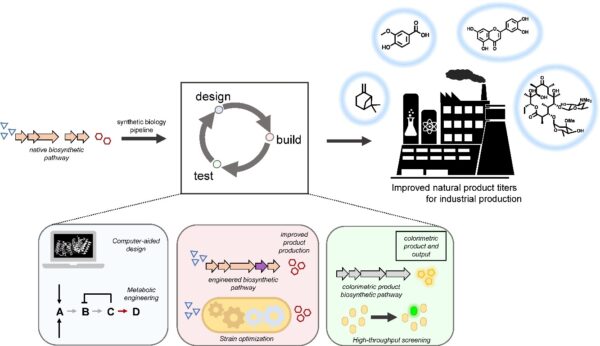
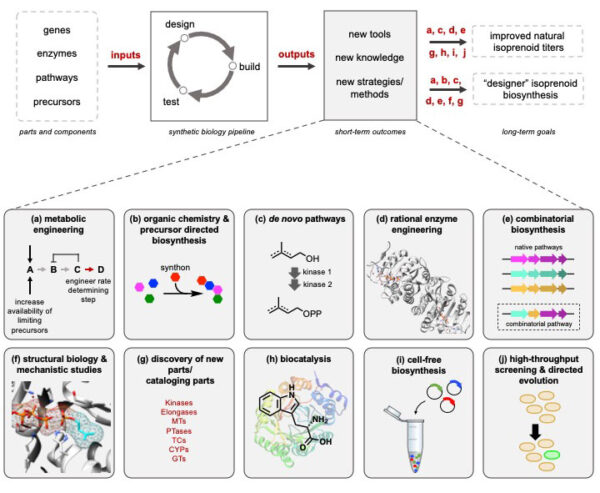
[49] Malico AA, Calzini MA, Gayen AK, Williams GJ*. “Synthetic biology, combinatorial biosynthesis, and chemo-enzymatic synthesis of isoprenoids“. 2020, J Ind Microbiol Biotechnol, https://doi.org/10.1007/s10295-020-02306-3
[48] Malico A, Nichols L, Williams GJ*. “Synthetic biology enabling access to designer polyketides“. 2020, Curr Opin Chem Biol, 58, 45-53.
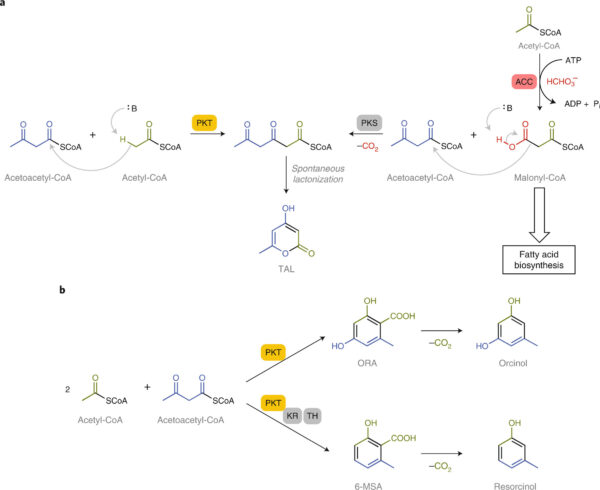
[47] Gayen A, Nichols L, Williams GJ*. “An artificial pathway for polyketide biosynthesis“. 2020, Nat Catalysis, 3, 536.
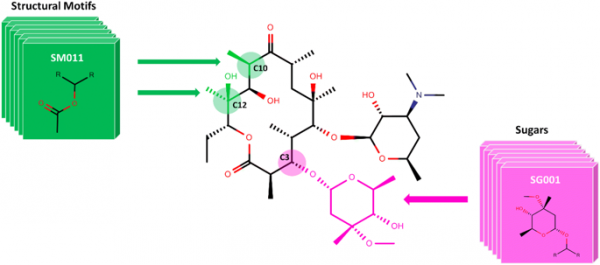
[46] Zin PP, Williams GJ, Fourches D*. “SIME: Synthetic Insight-based Macrolide Enumerator to Generate the V1B Library of 1 Billion Macrolides“, 2020, J Cheminform, 12, 23, https://doi.org/10.1186/s13321-020-00427-6
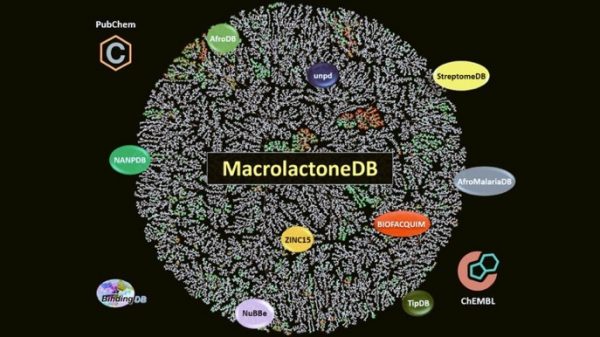
[45] Zin PP, Williams GJ, Ekins S*. “Cheminformatics Analysis and Modeling with MacrolactoneDB“, 2020, Scientific Reports, 10, 6284, https://doi.org/10.1038/s41598-020-63192-4
[44] Kalkreuter E, Bingham K, Keeler A, Lowell A, Schmidt J, Sherman DH, Williams GJ*. “Computationally-guided exchange of substrate selectivity motifs in a modular polyketide synthase acyltransferase”, 2020, bioRxiv 2020.04.23.058214
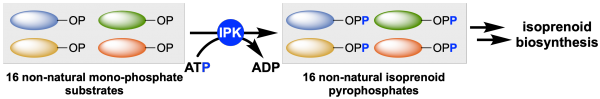
[43] Lund S, Courtney T, Williams GJ*. “Probing the substrate promiscuity of isopentenyl phosphate kinase as a platform for hemiterpene analogue production”, 2019, ChemBioChem, doi.org/10.1002/cbic.201900135.
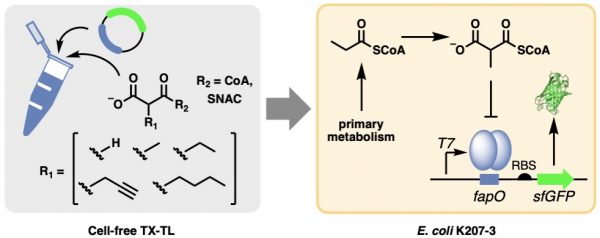
[42] Kalkreuter E, Keeler A, Malico A, Bingham K, Gayen A, Williams GJ*. “Development of a genetically-encoded biosensor for detection of polyketide synthase extender units in Escherichia coli”, 2019, ACS Synth Biol, 8, 6, 13891, doi/10.1021/acssynbio.9b00078.
[41] Lund S, Courtney T, Williams GJ*. “Probing the substrate promiscuity of isopentenyl phosphate kinase as a platform for hemiterpene analogue production”, 2019, ChemRxiv Preprint, .
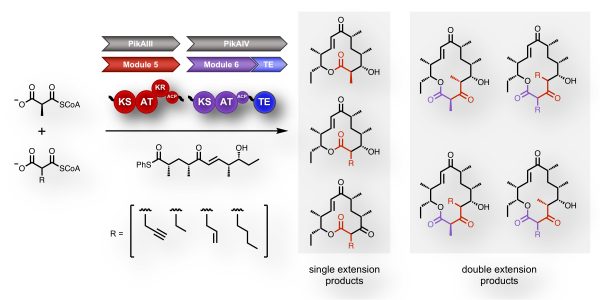
[40] Kalkreuter E, CroweTipton JM, Lowell AN, Sherman DH, Williams GJ*. “Engineering a modular polyketide synthase for sequential installation of unnatural extender unit substrates”, J Am Chem Soc, 2019, 141, 1961-1969.
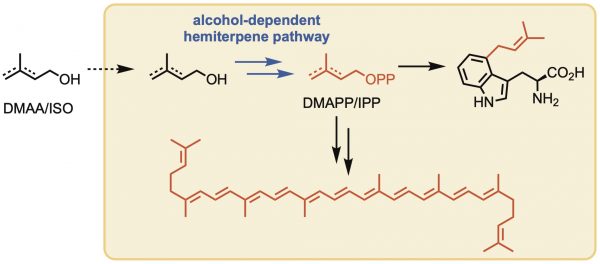
[39] Lund S, Hall R, Williams GJ. “Artificial pathway for isoprenoid biosynthesis decoupled from native hemiterpene metabolism“, ACS Synth Biol, 2019, 8, 2, 232-238.
“Most read” article (Jan-Feb) at ACS Synth Biol
Highlighted in Nature Catalysis, 2019, 2, 104
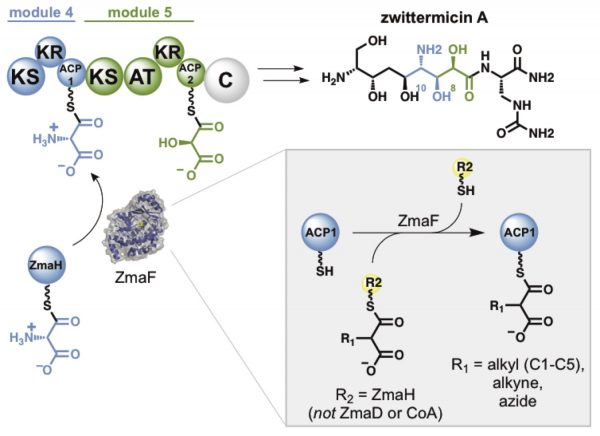
[38] Carpenter S, Williams GJ. “Extender unit promiscuity and orthogonal protein interactions of an aminomalonyl-ACP utilizing trans-acyltransferase from zwittermicin biosynthesis”, ACS Chem Biol, 2018, 13, 3361-3373.
[37] Zin PP, Williams GJ, Fourches D. “Cheminformatics-based Enumeration and Analysis of Large Libraries of Macrolides”. J Cheminformatics, 2018, online ahead of print.
[36] Carpenter SM, Kalkreuter E, Williams GJ. “Precursor-directed biosynthesis and semi-synthesis of natural products”. RSC Chemical Biology Series, Chemical and Biological Synthesis: Enabling Approaches for Understanding Biology, 2018, 275-312.
[35] Kasey C, Williams GJ. “Customizing Transcription-Factor Biosensors for Modern Biotechnology”. RSC Catalysis Series No. 32, Modern Biocatalysis: Advances Towards Synthetic Biological Systems, 2018, 205-233
[34] Kalkreuter E, Williams GJ. “Engineering Enzymatic Assembly Lines for the Production of New Antimicrobials”. Curr Opin Microbiol, 2018, 45, 140-148.
[33] Kasey C, Zerrad M, Li Y, Cropp TA, Williams GJ. “Development of Transcription Factor-Based Designer Macrolide Biosensors for Metabolic Engineering and Synthetic Biology“. ACS Synth Biol, 2017, 10.1021/acssynbio.7b00287.
“Most Read” Article (Nov-Dec) at ACS Synth Biol
[32] Nazari M, Malico A, Ekelof M, Lund S, Williams GJ, Muddiman, D. “Direct Analysis of Terpenes from Biological Buffer Systems using SESI and IR-MALDESI“. Anal Bioanal Chem, 2017, DOI:10.1007/s00216-017-0570-9.
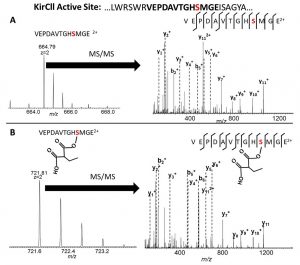
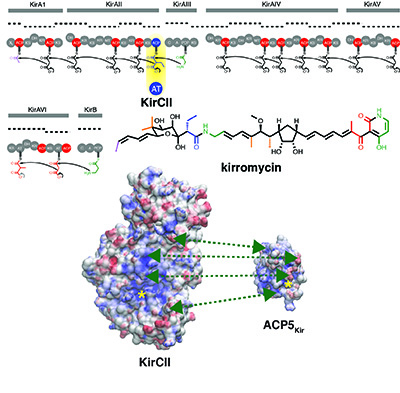
[31] Musiol-Kroll EM, Zubeil F, Schafhauser T, Härtner T, Kulik A, McArthur JB, Koryakina I, Wohlleben W, Grond S, Williams GJ, Lee SY, and Weber T. “Polyketide bio-derivatization using the promiscuous acyltransferase KirCII“. ACS Synth Biol, 2017, 6, 421.
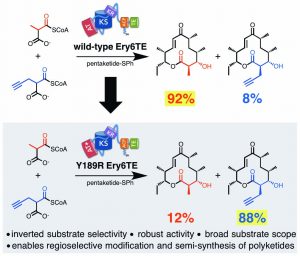
[30] Koryakina I, Kasey C, McArthur JB, Lowell AN, Chemler JA, Li S, Hansen DA, Sherman DH, and WilliamS GJ. “Inversion of Extender Unit Selectivity in the Erythromycin Polyketide Synthase by Acyltransferase Domain Engineering“. ACS Chem Biol, 2017, 12, 114.
Highlighted in Genetic Engineering & Biotechnology News
Highlighted in CEP Magazine, An American Institute of Chemical Engineers publication
NC State News Press Release
[29] Ladner C, Williams GJ. “Harnessing natural product assembly lines: structure, promiscuity, and engineering“. J Ind Microbial Biotechnol, 2016, 43, 371.
[28] Ye Z, Williams GJ. “Mapping a ketosynthase:acyl carrier protein binding interface via unnatural amino acid-mediated photocrosslinking“. Biochemistry, 2015, 53, 7494.
[27] Randall S, Koryakina I, Williams GJ, Muddiman DC. “Evaluating nonpolar surface area and LC-MS response: an application for site occupancy measurements for enzyme intermediates in polyketide biosynthesis”, Rapid Commun Mass Spectrom, 2014, 28, 2511.
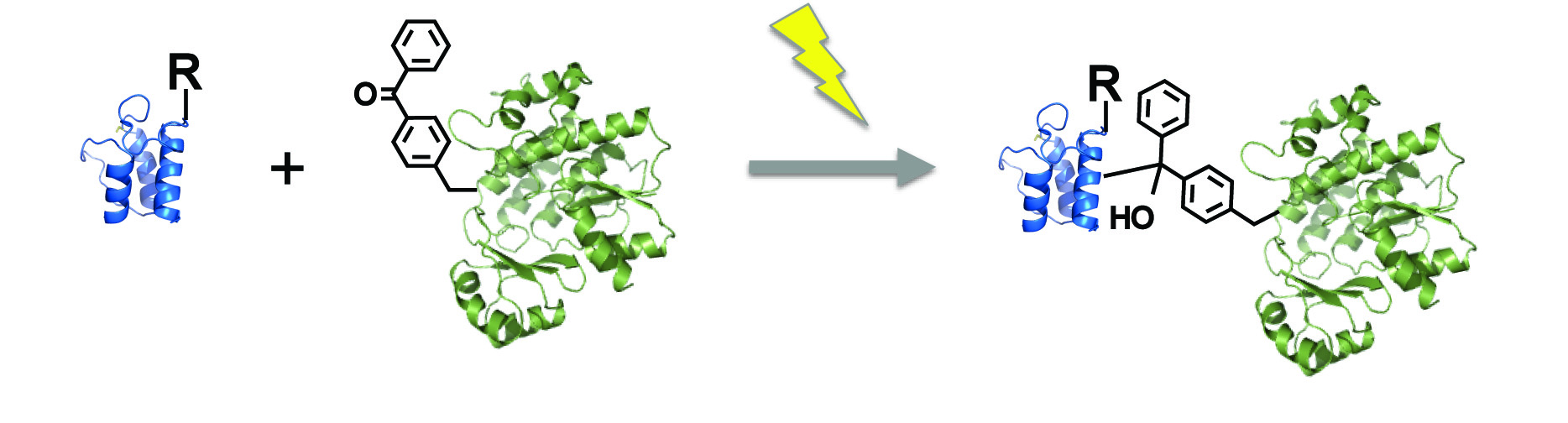
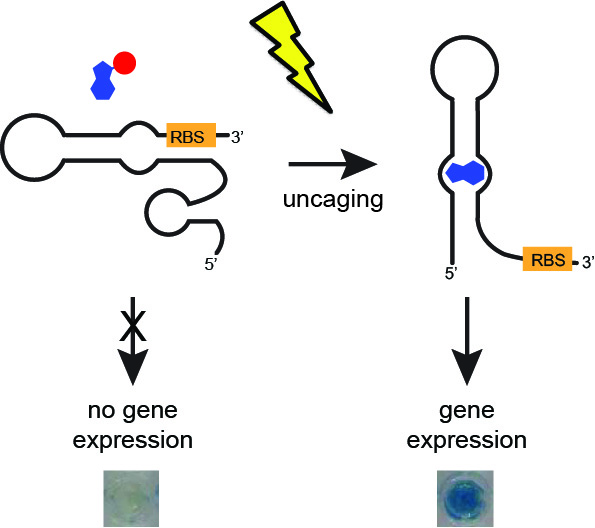
[26] Walsh S, Gardner L, Deiters A, Williams GJ. “Intracellular light-activation of riboswitch activity“. ChemBioChem, 2014, 15, 1346.
[25] Ye Z, Musiol EM, Weber T, Williams GJ. “Reprogramming acyl carrier protein interactions of an extender unit promiscuous trans-acyltransferase“. Chem Biol, 2014, 21, 636.
[24] Williams GJ. “Engineering polyketide synthases and non-ribosomal peptide synthetases”. Curr Opin Struct Biol, 2013, 23, 603. pdf
[23] Koryakina I, McArthur JB, Draelos, M, Randall S, Muddiman D, Williams GJ. “Reprogramming the biosynthesis of natural products by directed evolution”. Developments in Biotechnology and Bioprocessing, 2013, 9, 147-63.
[22] Koryakina I, McArthur JB, Draelos MD, Williams GJ. “Promiscuity of a Modular Polyketide Synthase Towards Natural and Non-Natural Extender Units”. Org Biomol Chem, 2013, 11, 4449-4458. pdf
 cover artwork in Org Biomol Chem
cover artwork in Org Biomol Chem
[21] Koryakina I, McArthur J, Randall S, Draelos MM, Musiol EM, Muddiman DC, Weber T, Williams GJ. “Poly specific trans-acyltransferase machinery revealed via engineered acyl-CoA synthetases”. ACS Chem Biol, 2013; 8, 200-208.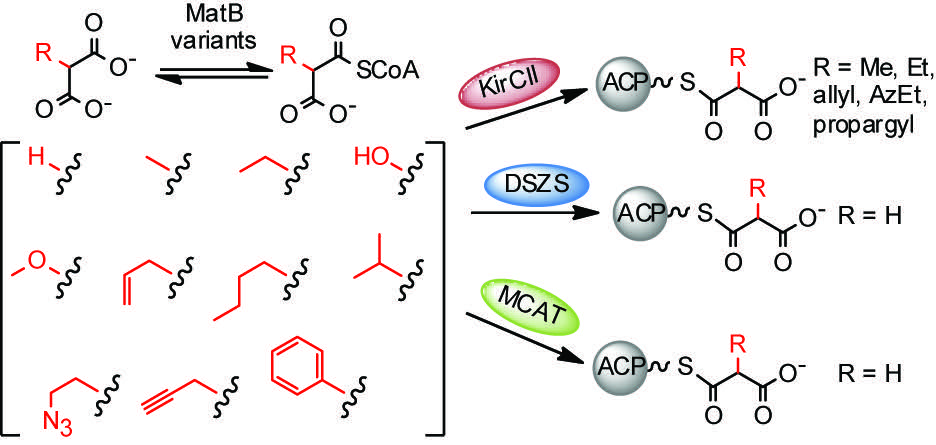
[20] McArthur J, & Williams GJ. “Engineering Glycosyltransferases”. In The Protein Engineering Handbook, Ed. Stefan Lutz, Wiley-VCH, Weinheim, 2012, Vol 3, 303-326.
[19] Ye Z, Desai H, Bair M, Williams GJ. “A novel photocrosslinking assay for reporting protein interactions in polyketide and fatty acid synthases”. Mol BioSys, 2011, 7, 3152-3156. pdf
[18] Koryakina I, Williams GJ. “Mutant malonyl-CoA synthetases for polyketide extender unit generation”. ChemBioChem, 2011, 12, 2289-2293. pdf
[17] Parajuli N, Williams GJ. “A high-throughput screen for directed evolution of aminocoumarin amide synthetases”. Anal Biochem, 2011; 419: 61-66. pdf
[16] Koryakina I, Neville J, Nonaka K, van Lanen S, Williams GJ. “A high-throughput screen for directed evolution of the natural product sulfotransferase LipB”. J Biomol Screening, 2011; 16: 845-851. pdf
Postdoctoral and graduate:
15. Williams GJ, Yang J, Zhang C, Thorson JS. “Recombinant E. coli prototype strains for in vivo glycorandomization”. ACS Chem Biol, 2011, 6, 95-100.
14. Williams GJ, Thorson JS. “Natural product glycosyltransferases: properties and applications. Adv Enzymol Relat Areas Mol Biol, 2009, 76, 55-119.
13. Williams GJ, Gantt RW, Thorson JS. “The impact of enzyme engineering upon natural product glycodiversification”. Curr Opin Chem Biol, 2008, 12, 556-564.
12. Gantt RW, Goff RD, Williams GJ, Thorson JS. “Probing the aglycone promiscuity of an engineered glycosyltransferase”. Angew Chemie Int Ed, 2008, 47, 8889-8892.
11. Williams GJ, Zhang C, Goff R, Thorson JS. “Optimization of a natural product glycosyltransferase for glycorandomization of novobiocin”. Chem Biol, 2008, 15, 393-401.
10. Williams GJ, Thorson JS. “A high-throughput fluorescence-based glycosyltransferase screen and its application in directed evolution”. Nat Protocols, 2008, 3, 357-362.
9. Williams GJ, Zhang C, Thorson JS. “Expanding the promiscuity of a natural product glycosyltransferase by directed evolution”. Nat Chem Biol, 2007, 3, 657-662.
8. Williams GJ, Woodhall T, Farnsworth LM, Nelson A, Berry A. “Stereochemically complementary biocatalysts created by directed evolution”. Synfacts 2007, 2, 0208-0208.
7. Williams GJ, Woodall T, Farnsworth LM, Nelson A, Berry A. “Creation of a pair of stereochemically complementary biocatalysts”. J Am Chem Soc, 2006, 128, 16238-47.
6. Williams GJ, Woodhall T, Nelson A, Berry A. “Structure-guided saturation mutagenesis of N-acetylneuraminic acid lyase for the synthesis of sialic acid mimetics”. PEDS 2005; 18: 239-246.
5. Woodhall T, Williams GJ, Berry A, Nelson A. “Synthesis of screening substrates for the directed evolution of sialic acid aldolase: Towards tailored enzymes for the preparation of influenza A inhibitor analogues”. Org Biomol Chem, 2005; 3: 1795-1800.
4. Woodhall T, Williams GJ, Berry A, Nelson A. “Creation of a tailored aldolase for the parallel synthesis of sialic acid mimetics”. Angew Chemie Int Ed, 2005; 44: 2109-2112.
3. Williams GJ, Nelson A, Berry A. “Directed evolution of enzymes for biocatalysis and the life sciences”. Cell Mol Life Sci, 2004; 61: 3034-3046.
2. Williams GJ, Berry A. “Directed evolution”. Biochemist 2003, 25, 4, 13-16.
1. Williams GJ, Doman S, Nelson A, Berry A. “Modifying the stereochemical course of an enzyme catalysed reaction by directed evolution”. Proc Natl Acad Sci USA, 2003, 100, 3143-3148.


 NC State Press Release
NC State Press Release